|
PROTOCOLS
and METHODS
when: pliocene and pleistocene
basket
5 - time-scales will be measured before 2000 alike before
present
Time is
timeless
(of course) hence
dealing with genetic (so early genesis at first) alike with biology
and geology or astronomy and oceanography or some other deep
investigation into heart of all these our immense living
bodies we cannot pretend to use as our time-scale a simple (unuseful) segment of the
whole time-scale (notwithstanding we don't know how
will be this whole)
in fact when
you do not have a simple but correct scale of time
(like in
our prehistory) there is simpler use the last
(so the first or the lowest) rung of the ladder: use Time Before Present
(BP) and
you are definitely correct
here we are
dealing with immense
distances
by Time:
the dendrochronology reach 10.400 perfect years ago and the Last
Glacial Maximum is suggested around 22.000 years ago when the Aurignacian prehistoric
culture is typical around 30.000 years ago and Neandertalers reach
80-90.000 100.000 years ago the Australopitecinae
are beyond 1.000.000 years elder and Pliocene
paradise is well beyond 3.000.000 years at least
honestly speaking
it is unwise deals with these
things and hear again and again: before and after Jesus Christ
before and after Communist before and after Fascist before
and after before and after..... who care?
before and
after
Rome or Buddah do not worth (and do not warrant)
a time-scale..... it appear that scholars investigating
the incredible
universe of chimpanzees and primates or of plancton and pollen
or of planets and comets are attempting to humanize the whole world jailing it in a frame they
can hold
safely: no more wide of that no more deep of that
no more long of that no more short of that no more
frozen of that no more wet of that no more dry
of that no more hot of that no more no more no
more..... that's the frame
but who
pretend to anthropomorphize Nature?

sixty years
ago they (of course they are ever) boasted that
Pleistocene was long one million
years (no more that was the distance they could manage
into their skull really their frame) but next appeared Robert Ardrey and
Elaine Morgan
who seemingly do not known nothing of Pliocene and Pleistocene and smashed
the frame.....
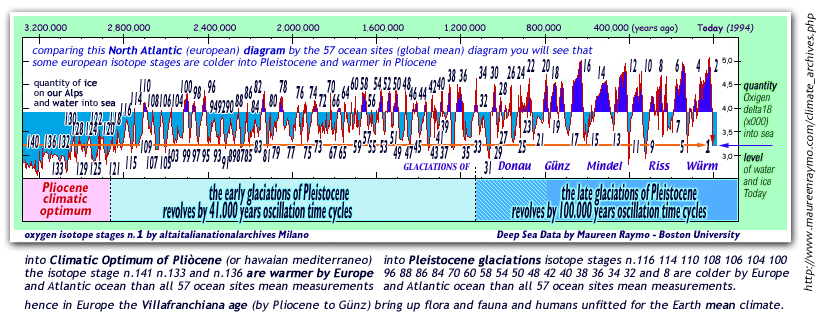
today diagrams
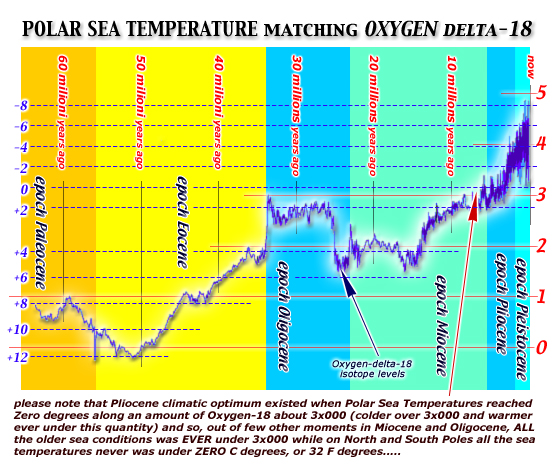
by oxigen-18 isotope stages (by Cesare Emiliani and Maureen
Raymo stratigraphies)
show us that Pleistocene was at least 2.600.000 years long
just in that
years prof. Colin
Renfrew
who knows well Pliocene and Pleistocene smashed other
distances (other totem and taboo also) showing that prehistory down untouchable
Europe
was (is) very very long according radiocarbon
and dendrochronology investigations
noteworthy
who pretend to use a framing
time-scale
sometime could be wrong according his scale: in the mediaeval
era was sure useful the chronology compiled by Dionisius "the
minus"
alike is useful again sunday morning into your church
mass (of course)
but when you have to count
years by whatsoever reason (like is when you keep
lectures by your University for instance) you
cannot use it or (for sure) you must not use it because
the whole count into this time-scale is wrong (by seven years
you know) no matter if seven years do not care at all..... it
simply is wrong and so (simply) you must do not use it
you do not
need any reason to be honest with TIME..... you simply
must be
[ Dionisius
diagram is on our BASIC PROTOCOL ] [ click to enlarge
]
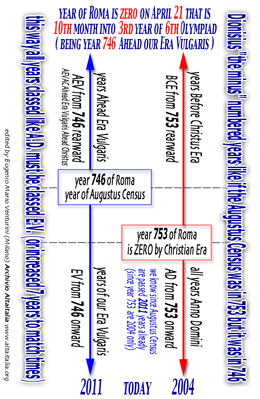
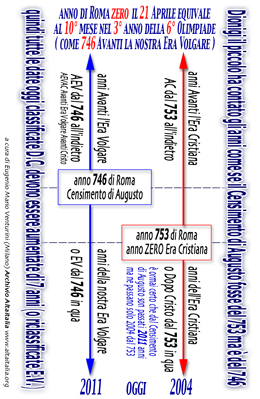
[ english read
this calendarius Dionisius ] [ italian
read this calendario Dionigi ]
nevertheless
a time-scale must describe TIME within readable limits (of course) but not
frame or bound it by arbitrary purposes: for sure a mediaeval
or liturgical time-scale will be unwise dealing
with ecosystems and chimpanzees or neandertalians as well
here we must
investigate TIME
of PLIOCENE
and PLEISTOCENE
at first [1] we must investigate
the early genesis of our KAPPA haplogroup hence all about the
stratified layers before birth of actual whole group so
investigating primates into Pliocene paradise (like the so-called Proconsul and
most recent Australopitecinae) down to last glaciation of Pleistocene (named Wurm) investigating
elder Heidelberg and Neander and Cro-magnon humans encompassing whole
Villafrancan until early Aurignacian periods or cultures (since
three or four millions years ago and up to 50.000-100.000 years
ago) over the floors or layers stratifications where next above
them will start our KAPPA group
that was the
early habitat and life of our prototype populations:
from this distant landscape we retain today a deep kinship
with water and his swimmers like
dolphins and whales..... and retain unforgettable words and rituals
as well

our fair
and wet ancestral
territory
here into pliocene
climatic optimum
(that is a typical
hawaian climate on mediterranean shores) was the same already
inherited by our most distant miocene's ancestors (like
is typical by Primates on whole Earth) maybe commencing on oxigen
isotope stage n.222 when the miocene hot climate turned fair into pliocene
with the arctic sea temperature that sink to zero degrees (that is 32 farenheit)
about 5.200.000 years ago and by 2.000.000 fantastic years onward
here is
a map of our pliocenic paradise
[ these
maps are on our BASIC PROTOCOL ] [ click for a better
view ]
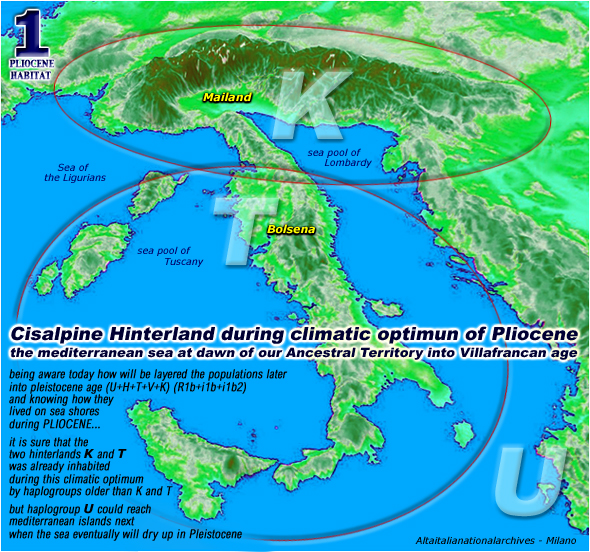
please note
that these ellipses are encircling the habitat of our genesis
even if haplogroups K and T will emerge later (about
60.000 years ago)
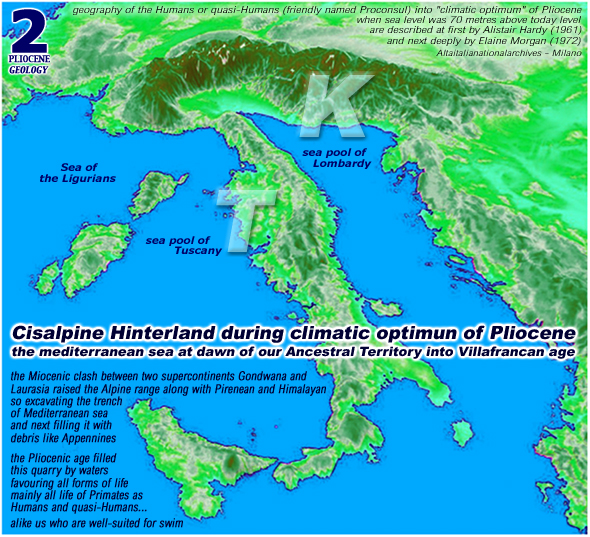
[ italian read
habitat del Pliocene ] [ italian
read geologia del Pliocene ]
slowly next
during many thousand years we began to experience some dry and
cold climatic long seasons revolving every 40.000 years
about (exactly 41.000) until their maximum turned immense
and very cold alike 100.000 frozen years (every round) became
typical
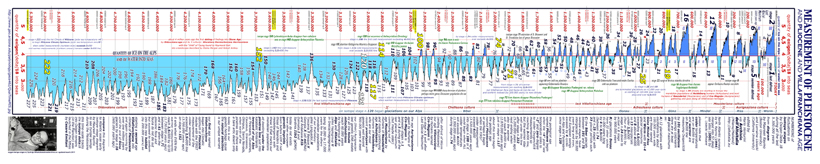
during a million
years at least twelve deep frozen waves (into five seasons
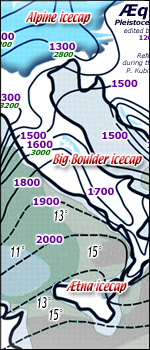
named Donau, Gunz, Mindel, Riss, and Wurm) seated on our Alps
and Pyreneans and Sierra Nevada and Cevennes and Big Boulder
and Olympus and Caucasus where the ice-cap reached 2.000 meters above the valley bottom
when all around there was the dust of arctic desert..... less
the leewardside of these ice-caps where the sinking katabatic
fair wind preserved year-round a very nice dwelling hinterland
just here atop
one of ultimate ice waves
about 60.000 years ago (if not many years before) appeared our KAPPA group out of our optimistic
ancient population
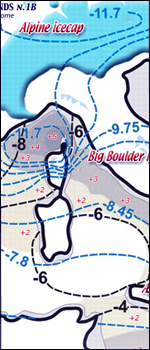
see two first
maps of frozen pleistocene reading aequilibrium line
altitude (ELA) and his temperature along with Mediterranean
sea surface temperatures (SST) during typical glaciations
like the Last
Glacial Maximum (LGM)
where figures show how dwelling hinterlands was more favourable
leeward side of icecaps (north or south) than otherwise.....
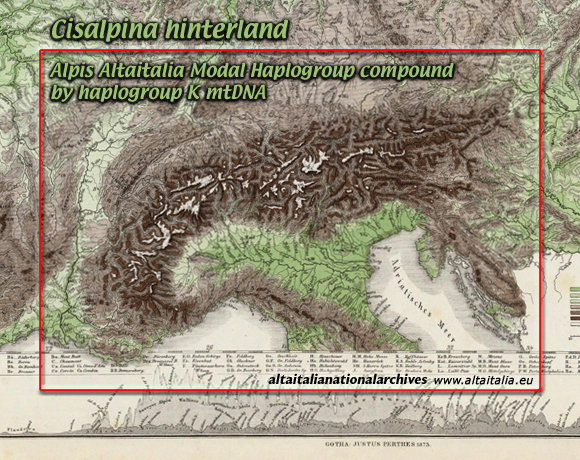
mainly on our alpine/cisalpine hinterland just on the
plain
of Lombardy
equilibrium
line altitudes
during last glacial maximum
[ maps and
diagram are on our BASIC PROTOCOL ] [ click to enlarge
]
[ italian
read equilibrio and temperatura durante L'ultimo Glaciale
Massimo and ELA diagram]
following [2] by the birth of our KAPPA haplogroup so completing the
genesis at his climax 50.000-60.000 years ago during the first
high wave of Wurm
and next going to the Last Glacial Maximum of Pleistocene (the
ultimate glacial maximum about 32.000-22.000 years ago) when
huge ice-cap got double thickness above our Alps
living
into Aurignacian prehistory seemingly to be highest point of palaeolithic
cultures
here our group dwelled
10.000 frozen years again
in those endless bright
seasons
maybe friendly groups joined our
alpine hinterland and meets our needs so matching our genetic
compound maybe forever or maybe
until defrost
10400-12400 years ago
when glaciers began melting and mammuths along with sabretooth
tigers and all reindeers slowly began moving north.....

nevertheless
we remained here
and
sure we all keep
contact
thru high alpine passes broken down by tonnes of melting
ice
in this new
season our ancient aurignacian calendars was re-designed
matching the newly
opened (free of ice-cap) distant horizons just when the four seasons
matched the perihelion between year 11.640 (-23:18 hours)
and 11.501 (+23:47 hours) during a whole century as our ancient proverbs remember
it again today
.....and next
the stone tools was abandoned for copper and brass
and iron spreading into the modern Villanovan culture until roughly 2.000
years ago
moreover [3] there will be possible
to track time since friendly peoples joined our Alpine and Altaitalia
Modal Haplogroup compound (AAMH) around the steady KAPPA haplogroup
nucleus .....but all useful dates will stop around 2.000 years
ago when
Roman Calendar superseded our ancient (former Aurignacian)
Villanovan Calendar of course
some other
researches dealing with younger times (from year zero
of our Era Vulgaris onward) are of genealogy interest and so
will be described into your genealogy pages
these diagrams
will be our basic time-scale
[ all diagrams
are on our BASIC PROTOCOL ] [ click to enlarge ]
[ english read
this oxygen-18 diagram ] [ italian
read this ossigeno-18 diagram ]
this first
TIME DIAGRAM show levels of Oxygen-delta18
during our last 65 million years: note that zero-degrees
into polar seas water was reached on pliocenic age only.....
when before PLIOCENE the polar seas water
was warmer than zero degrees ever
an unique climate
optimum
here under,
two diagrams show towering icecaps above Arctica and Antarctica,
Himalaia, Caucasus, Olympus, Gran Sasso, Cevennes, Pyreneans,
and Alpine mountains during our last 300.000 years
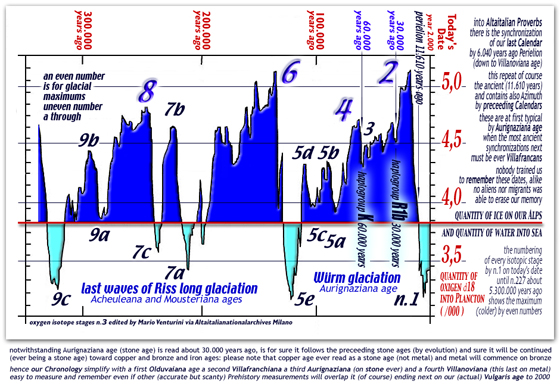
[ english read
this Wurm diagram ] [ italian
read this Wurm diagram ]
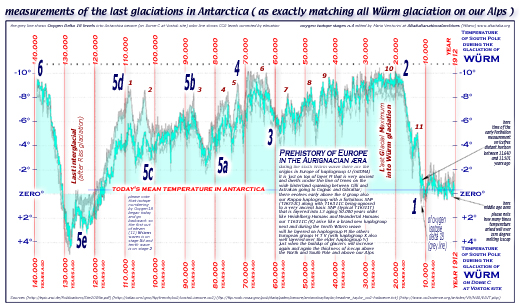
[ this antarctica
icecore diagram ] [ this
antartide icecore italian diagram ]
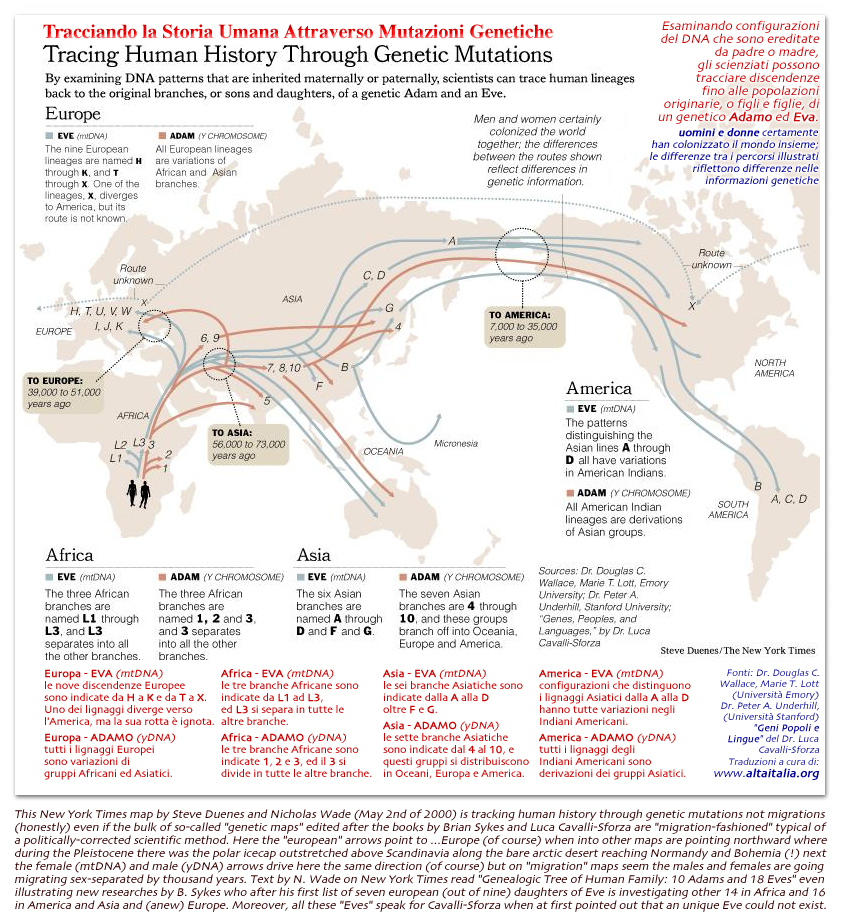
here at last
the Time will be measured by
genetic distance or time between SNP bases (like in the diagram n.1 by prof.
Cavalli-Sforza on this page) so locating all ancient events on
a readable time-scale and next matching the main events
accordingly by comparing other precision time-scales like radiocarbon
and dendrochronology and oxigen isotope stages
diagrams
that will be
useful with all screening results
NEXT
deeping into life
of KAPPA group
we think that genetic results can be compared by whatsoever
you wish to measure..... but we do not: here we
must compare only with data by fixing
date
for instance:
language elements will be compared only if could help to date something
(in a timeless past) alike fix something into environment where
otherwise some dating could be measured..... like words
embedded into proverb rhymes because are possible time-markers on the genesis
of proverb itself
alike if precision
dating will be our goal
.
|